-Search query
-Search result
Showing 1 - 50 of 87 items for (author: arita & k)

EMDB-28957: 
Cryo-EM structure of the Agrobacterium T-pilus
Method: helical / : Kreida S, Narita A, Johnson MD, Tocheva EI, Das A, Jensen GJ, Ghosal D

PDB-8fai: 
Cryo-EM structure of the Agrobacterium T-pilus
Method: helical / : Kreida S, Narita A, Johnson MD, Tocheva EI, Das A, Jensen GJ, Ghosal D

EMDB-17402: 
Uncharacterized Q8U0N8 protein from Pyrococcus furiosus
Method: single particle / : Pacesa M, Correia BE, Levy ED

EMDB-18415: 
Cysteine tRNA ligase homodimer
Method: single particle / : Pacesa M, Correia BE, Levy ED

PDB-8p49: 
Uncharacterized Q8U0N8 protein from Pyrococcus furiosus
Method: single particle / : Pacesa M, Correia BE, Levy ED

PDB-8qhp: 
Cysteine tRNA ligase homodimer
Method: single particle / : Pacesa M, Correia BE, Levy ED
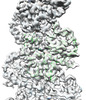
EMDB-33007: 
A CBg-ParM filament with ADP
Method: helical / : Koh A, Ali S, Robinson R, Narita A
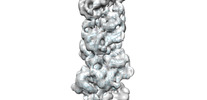
EMDB-33008: 
A CBg-ParM filament with GTP and a short incubation time
Method: helical / : Koh A, Ali S, Robinson R, Narita A
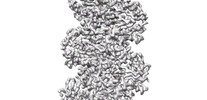
EMDB-33009: 
A CBg-ParM filament with ADP
Method: helical / : Koh A, Ali S, Robinson R, Narita A
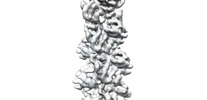
EMDB-33012: 
A CBg-ParM filament with GTP or GDPPi
Method: helical / : Koh A, Ali S, Robinson R, Narita A

PDB-7x54: 
A CBg-ParM filament with ADP
Method: helical / : Koh A, Ali S, Robinson R, Narita A

PDB-7x55: 
A CBg-ParM filament with GTP and a short incubation time
Method: helical / : Koh A, Ali S, Robinson R, Narita A

PDB-7x56: 
A CBg-ParM filament with ADP
Method: helical / : Koh A, Ali S, Robinson R, Narita A

PDB-7x59: 
A CBg-ParM filament with GTP or GDPPi
Method: helical / : Koh A, Ali S, Robinson R, Narita A

EMDB-33200: 
Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form)
Method: single particle / : Onoda H, Kikuchi A, Kori S, Yoshimi S, Yamagata A, Arita K
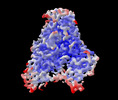
EMDB-33201: 
Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form)
Method: single particle / : Onoda H, Kikuchi A, Kori S, Yoshimi S, Yamagata A, Arita K
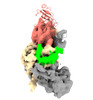
EMDB-33298: 
Cryo-EM map of apo-DNMT1 (aa:351-1616)
Method: single particle / : Onoda H, Kikuchi A, Kori S, Yoshimi S, Yamagata A, Matsuzawa S, Arita K
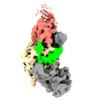
EMDB-33299: 
Cryo-EM map of DNMT1 (aa:351-1616) in complex with ubiquitinated H3
Method: single particle / : Onoda H, Kikuchi A, Kori S, Yoshimi S, Yamagata A, Arita K

PDB-7xi9: 
Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form)
Method: single particle / : Onoda H, Kikuchi A, Kori S, Yoshimi S, Yamagata A, Arita K

PDB-7xib: 
Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form)
Method: single particle / : Onoda H, Kikuchi A, Kori S, Yoshimi S, Yamagata A, Arita K

EMDB-14181: 
Human collided disome (di-ribosome) stalled on XBP1 mRNA
Method: single particle / : Denk TG, Tesina P, Beckmann R

PDB-7qvp: 
Human collided disome (di-ribosome) stalled on XBP1 mRNA
Method: single particle / : Denk TG, Tesina P, Beckmann R

EMDB-14153: 
SARS-CoV-2 Spike, C3 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

PDB-7qus: 
SARS-CoV-2 Spike, C3 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

EMDB-14152: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

EMDB-14154: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C1 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

EMDB-14155: 
SARS-CoV-2 Spike, C1 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

PDB-7qur: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

EMDB-32629: 
Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution
Method: helical / : Akil C, Ali S, Tran LT

EMDB-32630: 
Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution
Method: helical / : Akil C, Ali S, Tran LT

EMDB-30988: 
Structure of Enolase from Mycobacterium tuberculosis
Method: single particle / : Bose S, Vinothkumar KR

EMDB-30989: 
Structure of PEP bound Enolase from Mycobacterium tuberculosis
Method: single particle / : Bose S, Vinothkumar KR

PDB-7e4x: 
Structure of Enolase from Mycobacterium tuberculosis
Method: single particle / : Bose S, Vinothkumar KR

PDB-7e51: 
Structure of PEP bound Enolase from Mycobacterium tuberculosis
Method: single particle / : Bose S, Vinothkumar KR

EMDB-12178: 
Staphylococcus aureus 30S ribosomal subunit in presence of spermidine (body only)
Method: single particle / : Belinite M, Khusainov I, Marzi S, Romby P, Yusupov M, Hashem Y

EMDB-12179: 
Staphylococcus aureus 30S ribosomal subunit in presence of spermidine (head only)
Method: single particle / : Belinite M, Khusainov I, Marzi S, Romby P, Yusupov M, Hashem Y

PDB-7bgd: 
Staphylococcus aureus 30S ribosomal subunit in presence of spermidine (body only)
Method: single particle / : Belinite M, Khusainov I, Marzi S, Romby P, Yusupov M, Hashem Y

PDB-7bge: 
Staphylococcus aureus 30S ribosomal subunit in presence of spermidine (head only)
Method: single particle / : Belinite M, Khusainov I, Marzi S, Romby P, Yusupov M, Hashem Y

EMDB-12090: 
Staphylococcus aureus 30S ribosomal subunit in absence of spermidine with fixed helix 44
Method: single particle / : Belinite M, Khusainov I, Marzi S, Romby P, Yusupov M, Hashem Y

EMDB-12091: 
Staphylococcus aureus 30S ribosomal subunit in absence of spermidine with reoriented helix 44
Method: single particle / : Belinite M, Khusainov I, Marzi S, Romby P, Yusupov M, Hashem Y

EMDB-30636: 
Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment
Method: single particle / : Sakuragi T, Kanai R

PDB-7dce: 
Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment
Method: single particle / : Sakuragi T, Kanai R, Tsutsumi A, Narita H, Onishi E, Miyazaki T, Baba T, Nakagawa A, Kikkawa M, Toyoshima C, Nagata S

EMDB-30384: 
Simplified Alpha-Carboxysome, T=3
Method: single particle / : Tan YQ, Ali S, Xue B, Robinson RC, Narita A, Yew WS

EMDB-30385: 
Simplified Alpha-Carboxysome, T=4
Method: single particle / : Tan YQ, Ali S, Xue B, Robinson RC, Narita A, Yew WS

PDB-7ckb: 
Simplified Alpha-Carboxysome, T=3
Method: single particle / : Tan YQ, Ali S, Xue B, Robinson RC, Narita A, Yew WS

PDB-7ckc: 
Simplified Alpha-Carboxysome, T=4
Method: single particle / : Tan YQ, Ali S, Xue B, Robinson RC, Narita A, Yew WS

EMDB-23052: 
Staphylococcus aureus 30S ribosomal subunit in presence of spermidine
Method: single particle / : Belinite M, Khusainov I, Marzi S, Romby P, Yusupov M, Hashem Y

PDB-7kwg: 
Staphylococcus aureus 30S ribosomal subunit in presence of spermidine
Method: single particle / : Belinite M, Khusainov I, Marzi S, Romby P, Yusupov M, Hashem Y

EMDB-9757: 
Whole structure of a 15-stranded ParM filament from Clostridium botulinum
Method: helical / : Koh F, Narita A, Lee LJ, Tan YZ, Dandey VP, Tanaka K, Popp D, Robinson RC

EMDB-9758: 
Averaged strand structure of a 15-stranded ParM filament from Clostridium botulinum
Method: helical / : Koh F, Narita A, Lee LJ, Tan YZ, Dandey VP, Tanaka K, Popp D, Robinson RC
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
